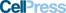Access provided by
Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors
Affiliations
- Laboratory of Molecular and Cell Biology, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
- Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Affiliations
- Laboratory of Molecular and Cell Biology, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
- Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Affiliations
- Laboratory of Molecular and Cell Biology, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
- Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Affiliations
- Laboratory of Molecular and Cell Biology, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
- Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Affiliations
- Razavi Newman Integrative Genomics and Bioinformatics Core, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Affiliations
- Laboratory of Molecular and Cell Biology, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
- Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Correspondence
- Corresponding author
 Correspondence information about the author Patrick D. Hsu
Correspondence information about the author Patrick D. HsuAffiliations
- Laboratory of Molecular and Cell Biology, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
- Helmsley Center for Genomic Medicine, Salk Institute for Biological Studies, 10010 N. Torrey Pines Road, La Jolla, CA 92037, USA
Correspondence
- Corresponding author
 Article Info
Article Info
Figure 1
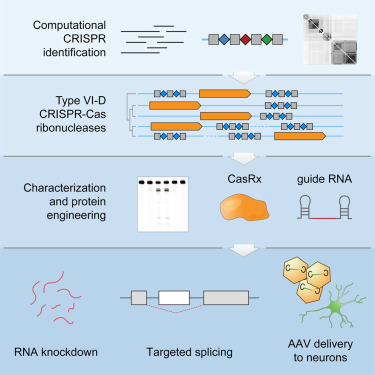
Type VI CRISPR-Cas13d Is a Family of Single-Effector CRISPR Ribonucleases
(A) Maximum-likelihood phylogenetic tree of Cas13d effectors used in this study, with the full Cas13d CRISPR locus depicted along with conserved HEPN RNase domains. Grey rectangles denote CRISPR direct repeats (DRs) and blue diamonds indicate spacer sequences.
(B) RNA sequencing of a heterologously expressed Cas13d locus from an uncultured Ruminococcus sp. sample. Mature gRNAs mapping to the CRISPR array indicate a processed 30 nt DR and a variable spacer length from 14–26 nt. Co-fold analysis of direct repeat truncation indicates a strong hairpin structure.
(C) Purified E. siraeum Cas13d and catalytically dead Cas13d (dCas13d) protein are each sufficient to process a guide array into its two component gRNAs. Addition of EDTA does not impair gRNA processing. “d,” dCas13d(R295A, H300A, R849A, H854A).
See also Figures S1–S3 and Table S1 .
Figure 2
Programmable RNA Targeting by Cas13d In Vitro
(A) E. siraeum Cas13d requires a matching guide array or mature gRNA to efficiently cleave complementary ssRNA targets. Denaturing gel depicts cleavage reactions incubated at 37°C for 1 hr. NT, non-targeting.
(B) Substitution with dCas13d or addition of EDTA abrogate Cas13d-mediated RNA targeting with both the guide and array. “d,” dead Cas13d.
(C) Denaturing gel depicting guide-target match-dependent activation of Cas13d cleavage activity. Scrambled target RNA (“A”) is fluorescently labeled, while guide-complementary activator target RNA (“B”) is unlabeled. RNA cleavage activity is abolished by the individual removal of guide RNA or complementary target RNA, as well as the addition of EDTA or the catalytic inactivation of Cas13d (indicated as “d”).
(D) A model for guide and target-dependent activation of Cas13d RNase activity. The ternary Cas13d:gRNA:target RNA complex is capable of cleaving the complementary target RNA or bystander RNAs.
See also Figure S4 and Table S1 .
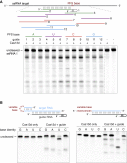
Figure 3
Characterization of Cas13d Target Substrate Preference
(A) Cas13d can be generalizably reprogrammed with multiple guides and does not exhibit a protospacer flanking sequence (PFS) requirement. RNA cleavage by EsCas13d and 12 guides tiling the target RNA is shown. Control lanes are from a separate gel run in parallel.
(B) Cas13d preferentially cleaves uracil bases in the loop of a hairpin or a linear homopolymer repeat, which is interrupted every 5 nt by a transition mutation (X) to enable synthesis.
See also Figure S4 and Table S1 .

Figure 4
RNA Knockdown Activity Screen of Engineered Cas13d Orthologs in Human Cells
(A) Schematic for mammalian expression constructs encoding for engineered Cas13d effectors and guides. NLS, nuclear localization signal. pre-gRNA, artificial unprocessed guide RNA containing a single 30 nt spacer sequence flanked by 2 full-length 36 nt DRs. gRNA, predicted mature guide RNA with a single 30 nt processed DR and 22 nt spacer sequence.
(B) Heatmap of mCherry protein knockdown in a Cas13d ortholog activity screen in HEK293FT cells using pools of 4 pre-gRNAs or gRNAs. Normalized MFI, median fluorescent intensity relative to non-targeting condition. Positions in gray were not tested, with n = 3.
(C) Immunocytochemistry of Cas13d showing localization and expression of engineered constructs. Scale bar, 10 μm. Blue pseudocolor, DAPI staining of nuclei.
(D) Comparison of Adm and Rfx Cas13d ortholog constructs for knockdown of endogenous B4GALNT1 mRNA reveals RfxCas13d-NLS (CasRx) to be most effective for both guide RNA architectures. Pools of 4 guides were used for targeting. NT, non-targeting. Values are mean ± SEM with n = 3.
See also Tables S2 , S4 , and S5 .
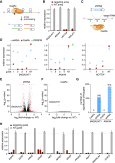
Figure 5
CasRx Mediates Efficient and Specific Knockdown of Diverse Human Coding and Noncoding Transcripts
(A) Multiple guide RNAs tiling a target transcript can be expressed as a single array and processed by RfxCas13d-NLS (CasRx) into individual gRNAs within the same cell.
(B) Arrays of 4 guides each mediate target knockdown by CasRx in 293FT cells via transient transfection. Knockdown relative to GFP vehicle control was determined by qPCR. Values shown as mean ± SEM with n = 3.
(C) Schematic of CasRx target sequences and spacer position-matched shRNAs.
(D) Relative target RNA knockdown by individual position-matched shRNAs and CasRx gRNAs. NT, non-targeting. CRISPRi, dCas9-mediated transcriptional repression. Values shown as mean ± SEM with n = 3.
(E) Volcano plot of differential transcript levels between B4GALNT1 targeting and non-targeting (NT) shRNAs as determined by RNA sequencing (n = 3). 542 non-specific transcript changes were identified.
(F) Volcano plot of differential transcript levels between B4GALNT1-targeting CasRx and non-targeting (NT) guide. Targeting guide position is matched to the shRNA shown in (E). B4GALNT1 was the only transcript exhibiting a significant change, with n = 3.
(G) Summary of significant off-target transcript perturbations by matched shRNAs and CasRx guides.
(H) CasRx targeting of 11 endogenous transcripts, each with 3 guides and a non-targeting (NT) guide in 293FT cells. Transcript levels are relative to GFP vehicle control, mean ± SEM with n = 3.
See also Figures S5 and S6 and Tables S2 , S3 , S4 , and S5 .

Figure 6
AAV Delivery of Catalytically Inactive dCasRx Splice Effectors to Manipulate Alternative Splicing
(A) Schematic of bichromatic exon skipping reporter. +1 and +3, reading frame; BP, intronic branch point-targeting guide; SA, splice acceptor site-overlapping guide; EX, exonic guide; SD, splice donor site-overlapping guide; AUG, start codon; UGA, stop codon. Inclusion of the second exon leads to an out-of-frame (+3), non-fluorescent translation of dsRed followed by in-frame mTagBFP2. Exclusion of the targeted exon leads to an in frame translation of dsRed (+1) followed by a stop codon.
(B) Induced exon exclusion by dCasRx and an N-terminal hnRNPa1-dCasRx fusion protein targeted to pre-mRNA. The Gly-rich C-terminal domain of hnRNPa1 is used as the effector domain. Exon skipping efficiency is depicted as a relative percentage of cells carrying primarily the dsRed or BFP isoform, determined through flow cytometry. ‘A’, CRISPR array carrying all 4 guides. Values are mean ± SEM with n = 3.
(C) AAV design carrying dCasRx and a three-guide array with total transgene size <4.3 kb, including AAV inverted terminal repeats (ITRs).
(D) Schematic of frontotemporal dementia (FTD) disease modeling. Neurons are generated via Neurogenin-2 (Ngn2) directed differentiation of patient-derived and control iPSCs followed by transduction with dCasRx or vehicle control AAV (EFS-mTagBFP2).
(E) FTD is associated with SNPs in a putative intronic splice enhancer following exon 10 of the MAPT transcript encoding for tau. Alternative splicing of MAPT exon 10 results in 4R tau (by inclusion) and 3R tau (by exclusion). SNPs in the intronic splice enhancer including the indicated IVS 10 + 16 mutation result in increased exon inclusion and higher levels of 4R tau. gRNAs contained in a dCasRx array were targeted to the exon 10 splice acceptor (g1) as well as two putative exonic splice enhancers indicated in purple (g2, g3).
(F) Relative 4R/3R tau transcript ratios in differentiated neurons were assayed via qPCR at 14 days following transduction with AAV. FTD, frontotemporal dementia cells carrying IVS 10 + 16. Values are mean ± SD with n = 3. ∗∗∗∗p < 0.0001.
See also Figure S5 and Tables S2 , S4 , and S5 .

Figure S1
Bioinformatic Pipeline for the Identification of the RNA-Targeting Class 2 CRISPR System Cas13d, Related to Figure 1
(A) Schematic describing a computational pipeline for CRISPR system identification. A minimal definition for a putative class 2 CRISPR locus was used, requiring only a CRISPR repeat array and a nearby protein > 750 aa in length. As per Methods, initial search was performed on prokaryotic genome assemblies derived from NCBI Genome, and later expanded via TBLASTN of predicted Cas13d proteins against public metagenome sequences without predicted open reading frames. DR, direct repeat.
(B) Phylogenetic classification and alignment of full-length Cas13d effectors and metagenomic fragments. Cas13d effectors and metagenomic Cas13d protein fragments cluster into several distinct branches, which are colored for ease of interpretation. Shading indicates residue conservation using the Blosum62 matrix. Full-length Cas13d effectors used in this study were sampled from distinct branches of the Cas13d family. Alignment of Cas13d proteins and protein fragments was performed using ClustalOmega 1.2.4 and maximum-likelihood tree building was performed with PhyML 3.2.
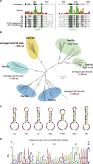
Figure S2
Phylogenetic Classification of RNA-Targeting Class 2 CRISPR Effectors and Sequence Conservation within the Cas13d Family, Related to Figure 1
(A) HEPN motif conservation in Cas13d effectors used in this study with conserved residues shaded according to Blosum62. The RxxxxH HEPN motif is highlighted.
(B) Maximum-likelihood tree of type VI CRISPR-Cas families. Average amino acid lengths of Type VI Cas13 superfamily effectors are indicated in red. Alignment of previously described class 2 CRISPR RNA-targeting proteins ( Abudayyeh et al., 2017; Cox et al., 2017; East-Seletsky et al., 2016, 2017; Smargon et al., 2017 ) and Cas13d effectors was performed using MAFFT 7.38 and maximum-likelihood tree building was performed with PhyML 3.2. Branch labels and scale bar indicate substitutions per site.
(C) Predicted Cas13d direct repeat RNA secondary structure.
(D) Sequence logo of full length 36 nt Cas13d direct repeats.
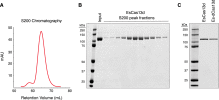
Figure S3
Purification of Recombinant Cas13d Protein, Related to Figure 1
EsCas13d was expressed as N-terminal His-MBP fusion and purified by successive affinity, cation exchange, and size exclusion chromatography. The His-tag was removed by TEV protease cleavage.
(A) Chromatogram from Superdex 200 column for EsCas13d.
(B) SDS-PAGE gel of size exclusion chromatography fractions for E. siraeum Cas13d.
(C) SDS-PAGE gel of purified E. siraeum Cas13d and dCas13d (R295A, H300A, R849A, H854A mutations of predicted catalytic residues in both HEPN motifs).
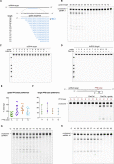
Figure S4
In Vitro Characterization of Cas13d Properties, Related to Figures 2 and 3
(A) Schematic showing the length and sequence of gRNA spacer truncations and spacer position relative to the complementary ssRNA target.
(B) Denaturing gel depicting EsCas13d cleavage activity of target RNA with different gRNA spacer lengths.
(C) Denaturing gel depicting EsCas13d cleavage reactions paired with 12 guides from Figure 3 A tiling a complementary ssDNA version of the ssRNA target.
(D) Denaturing gel depicting cleavage reactions using EsCas13d paired with the same 12 guides tiling a dsDNA version of the complementary target.
(E) Quantification of cleavage efficiency from Figure 3 A. Each PFS base is the average of 3 different spacer sequences tiling a complementary target RNA. Cleavage percentage is determined by the ratio of cleaved band intensity divided by total lane intensity. Mean is depicted ± SD with each data point representing an independent replicate.
(F) Cas13d-mediated cleavage of target RNA carrying different PFS bases given an invariant spacer sequence. Quantification of Cas13d cleavage efficiency and a representative denaturing gel depicting EsCas13d cleavage activity are shown. Differences are not significant (one-way ANOVA, p = 0.768). Cleavage percentage is determined as above, and mean is depicted ± SD with n = 3.
(G and H) Optimal temperature range for Cas13d activity. Denaturing gels depicting EsCas13d cleavage activity at temperatures ranging from 16-62°C for two different target RNAs.
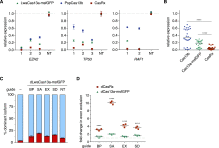
Figure S5
Comparison of Engineered Cas13 Superfamily Effectors for Targeted Knockdown and Splicing, Related to Figures 5 and 6
(A) Relative target RNA knockdown by individual position-matched gRNAs for CasRx, NLS-LwaCas13a-msfGFP ( Abudayyeh et al., 2017 ) and PspCas13b-NES ( Cox et al., 2017 ) in HEK293FT cells. NT, non-targeting. Values are mean ± SEM with n = 3.
(B) Comparison of Cas13 median knockdown efficiencies. n = 3 per guide RNA. ∗∗∗∗ indicates p < 0.0001 according to Friedman’s test.
(C) Exon exclusion by catalytically inactive NLS-dCas13a-msfGFP on the bichromatic splicing reporter. Guides are position-matched to those reported in Figure 6 B for CasRx. Values are mean ± SEM with n = 3.
(D) Comparison of splicing modulation by NLS-dCas13a-msfGFP and CasRx. Fold change in targeted exon exclusion relative to non-targeting guide is shown. Values are mean ± SEM with n = 3. ∗∗∗∗ indicates p < 0.0001 according to two-way ANOVA.

Figure S6
RNA Sequencing from CasRx and shRNA Targeting of ANXA4 in Human Cells, Related to Figure 5
(A) Volcano plots of differential transcript levels between ANXA4 targeting and non-targeting (NT) shRNAs as determined by RNA sequencing (n = 3). 915 non-specific transcript changes were identified.
(B) Volcano plot of differential transcript levels for an ANXA4 targeting CasRx array used in Figure 5 B containing a guide position matched to the shRNA shown in (A) and a non-targeting (NT) array. ANXA4 was the only transcript exhibiting significant downregulation with n = 3. HIST2HBE was the only transcript identified to exhibit significant upregulation. H2B is a dimer partner of H2AX ( Du et al., 2006 ) which has been shown to interact with ANXA4 ( Yang et al., 2010 ).
Highlights
- •Computational pipeline identifies the RNA-targeting type VI-D CRISPR-Cas family
- •Ortholog screen and protein engineering yields the programmable ribonuclease CasRx
- •CasRx RNA knockdown exhibits favorable efficiency and specificity relative to RNAi
- •Neuronal AAV delivery of dCasRx splice effectors alleviates tau mis-splicing
Summary
Class 2 CRISPR-Cas systems endow microbes with diverse mechanisms for adaptive immunity. Here, we analyzed prokaryotic genome and metagenome sequences to identify an uncharacterized family of RNA-guided, RNA-targeting CRISPR systems that we classify as type VI-D. Biochemical characterization and protein engineering of seven distinct orthologs generated a ribonuclease effector derived from Ruminococcus flavefaciens XPD3002 (CasRx) with robust activity in human cells. CasRx-mediated knockdown exhibits high efficiency and specificity relative to RNA interference across diverse endogenous transcripts. As one of the most compact single-effector Cas enzymes, CasRx can also be flexibly packaged into adeno-associated virus. We target virally encoded, catalytically inactive CasRx to cis elements of pre-mRNA to manipulate alternative splicing, alleviating dysregulated tau isoform ratios in a neuronal model of frontotemporal dementia. Our results present CasRx as a programmable RNA-binding module for efficient targeting of cellular RNA, enabling a general platform for transcriptome engineering and future therapeutic development.
Keywords:
CRISPR, genome engineering, gene editing, RNA targeting, RNA interference, alternative splicing, frontotemporal dementia, tau, Cas13, CasRxTo access this article, please choose from the options below
Read-It-Now
Now available: purchase access to all research journal HTML articles for 6 or 36 hours. Click here to explore this opportunity.
Purchase Access to this Article
Claim Access
If you are a current subscriber with Society Membership or an Account Number, claim your access now.
Subscribe to this Journal
Purchase a subscription to gain access to this and all other articles in this journal.